Introduction
Proteins are polymers of amino acids and 20 different amino acids arranged in infinite patterns to form different types of proteins. Proteins are involved in different roles in the living organisms, from carrying out important cellular functions like metabolic reactions to being an important structural component of animals, human and plant body parts.
Proteins studies in terms of their structure and functions and with increasing knowledge, it is concluded that the function of a protein is very much related to their structure. So, protein structural studies are very important in order to understand their functions. Proteins structure is resolved on different levels and terminology was assigned in order to understand the level of protein structure.
There are three basic levels of structure arrangement of a protein which consist of a single polypeptide, called primary protein structure, secondary protein structure, and tertiary protein structure. The primary protein structure is a simple sequence of the amino acids in which they arrange in a polypeptide chain.
The secondary structure of a protein is due to the folding of the polypeptide chain into different folds due to hydrogen bonding and Vander Waal forces. Whereas the tertiary structure of proteins is defined as the arrangement of secondary structure content in 3-dimensional space.
While some proteins consist of more than one polypeptide, their structure arranges into another level which is called quaternary structure due to the interaction between two or more polypeptides of that protein.
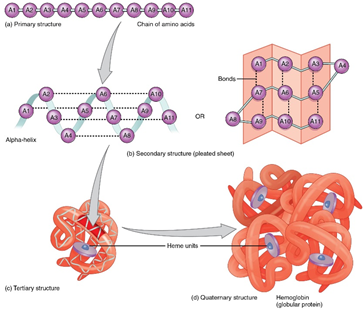
Fig. 1: The primary, secondary and tertiary structure of protein.
- Secondary structure of protein
The primary structure is very important in defining the structure and function of the protein. Amino acids join each other thorough peptide bonds which are rigid i.e., they do not allow rotation of the two amino acids freely. But the alpha carbon which is bond with NH- and C=O group have some rotation which allows arranging amino acids in different angles in limited values.
These angles are called torsion angles and help in the folding of the polypeptide chain into different secondary structure elements like α-helix, β-sheet, β pleated-sheet, and turns. These secondary structure elements are also stabilized by the forces present between amino acids located at some distance from each other.
These forces are hydrogen bonding and the van der Waal forces. Secondary structure elements present in repetitive forms in a protein and some proteins rich in α-helix content and others in β-sheet while others have mixed ratio of α-helix and β-sheet contents.
2.1 α-helix
The helical structure in the protein is one of the common secondary structure exist. There are different types of helical structure were observed in the proteins but the most common is the α-helix. The helical structure forms due to the presence of the turns in the polypeptide chain and different helical structure are identified on the basis of the number of amino acids a turn contains and the rise of the helical structure along its axis per turn.
The helical structure was first proposed in 1930 by William Astbury, but his description of the α-helix was later proved wrong. In 1952, a team of three scientists Linus Pauling, Robert Corey, and Herman Branson described the α-helix andβ-sheet structures in somewhat detail and with the correct description.
They also showed that the α-helical structure in nature has handedness that the polypeptide chain either turn in the clockwise (right-handed) or anticlockwise (left-handed) manner. The right-handed α-helical structure occurrences are the most common among the protein structures. The X-ray diffraction structure of the myoglobin was resolved in 1960 which confirmed the finding of the Pauling, Corey, and Branson and the right-handed α-helical structure was commonly found in myoglobin.
The α-helical structure is stabilized by the presence of the hydrogen bond formed between the peptide carbonyl group (C=O) and the peptide amide group (N-H) of the amino acid which is present four residues away. Each turn of the α-helix contains 3.6 amino acids and the helical structure rise along its axis to 5.4 Å.
The helical structure in most of the protein consisting of 12 amino acids but in some cases, helical stretch consists of 50 residues. The backbone of the polypeptide chain in the α-helical structure is present towards the inside, whereas R – group is pointed outwards of the α-helix. Due to the outward positioning of the R-group, any steric hindrance is avoided.
The α-helical structure is further stabilized by the presence of the van der Waal forces results in the tightly packed structure. the α-helical structure is most commonly found in membrane proteins as the backbone of a polypeptide is hydrophilic present inside of the structure, whereas R-group of the hydrophobic amino acids presents outwards which can easily interact with the hydrophobic environment of the membranes.
2.2 β pleated-sheet
β pleated-sheet is another most commonly found secondary structure in the proteins. β pleated-sheet structure consists of the stretched of adjacent polypeptide chains formed by the hydrogen bonding between the adjacent polypeptide chains. The polypeptide chains arranged in the same direction are called parallel β pleated-sheet and if they are arranged in the opposite direction are called anti-parallel β pleated-sheet.
2.2.1 Parallel β pleated-sheet
In the parallel β pleated-sheet adjacent polypeptide chains run in the same direction it means that the N- of all the polypeptide chains present at the same direction as their C-terminal present in the same direction. The parallel β pleated-sheet are rarely present as the secondary structure element and they are also less stable than anti-parallel β pleated-sheet because the hydrogen bonds form between adjacent polypeptide chains are longer and their conformation is unfavorable making them weaker.
2.2.2 Anti-parallel β pleated-sheet
In the anti-parallel β pleated-sheet, the adjacent polypeptide chains run in the opposite direction which means that the N-terminal region of one polypeptide chain and C-terminal region of the other polypeptide chain in the same direction.
This structure is the most commonly found β pleated-sheet secondary structure in the proteins. It is a more stable structure than the parallel β pleated-sheet because the hydrogen bond is more straight due to this distance of the bond is smaller making it stronger bonding.
The structure of the β pleated-sheet was also first identified the William Astbury in the 1930s but again his description of the β pleated-sheet structure does not meet new structural findings because of the unavailability of the necessary bonding data. Pauling and Corey, in 1952 along with α-helix structure description had defined β pleated-sheet correctly. Several proteins contain a mixed parallel and anti-parallel β pleated-sheet structure.
The structure appears sheet-like because of the zig-zag shape which is due to the α-carbon of one amino acid residue that appears at the top and it adjacent residue α-carbon place in the bottom in a repetitive manner, whereas R-group are stretched outwards. The distance between the two adjacent amino acids is 7 Å and on average one strand in β pleated-sheet contains 6 amino acid residues and in several cases up to 15 residues. The sheet has a slight helical turn due to maintenance of conformational stability within the chains which is caused by the hydrogen bonding between adjacent polypeptide chains. This turn is right-handed in nature.
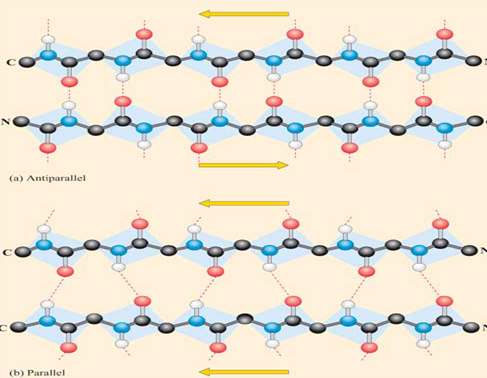
Figure 2: showing the β-pleated sheet structure.
A β pleated-sheet can consist of 6 polypeptide strands on average and in several cases, there are 15 strands present in a sheet. In a β pleated-sheet, hydrogen bonding can be between the strands of a polypeptide line up adjacent to each other which are formed due to the turns at a sharp angle.
Mostly, proline residue is present in these turn and they are called β turn. In other cases, polypeptide strands located at different places in a protein can form a hydrogen bond with each other and these are often joined by a long stretch of a polypeptide called loops and sometimes secondary structure like α-helix present in loop regions. The turn of the loop region which joined the two strands can be a right-handed cross over or a left-handed cross over which is rarely present in a β pleated-sheet.
2.3 Loop structure
The third secondary structure which presents in the protein is the loop structure which joins the other secondary structure such as α-helix and strands of β-sheet. The loop structure consists of 2-6 amino acids. As mentioned above the secondary structure element arrangement in 3-dimensional space gives the shape to the protein. In many cases, the arrangement of protein in 3 dimension space requires a change in direction of the polypeptide chain and these loop regions are present in such places to turn the polypeptide chain in a specific direction.
The most common type of loop region present in a protein is β-turn which consists of 4 amino acids and help in joining the adjacent strand of a β- pleated sheet. β-turn is stabilized by the formation of the hydrogen bond between the carbonyl group (C=O) of the first amino acid and the amide group (N-H) of the fourth amino acids. Proline is commonly present in such a turn because its structure provides the necessary bend to the turn. β-turn type I and type II differs based on the difference in the torsion angles.
Another type of loop structure present in the protein is called the omega loop which consists of 6 amino acids residue. It is a compact structure and because it attains the shape of the Greek word (Ω) hence given the name omega loop. These loop structures are mostly present on the surface of the protein where they help in the recognition role.
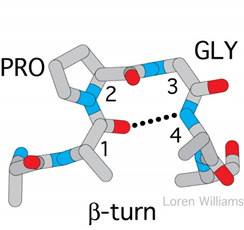
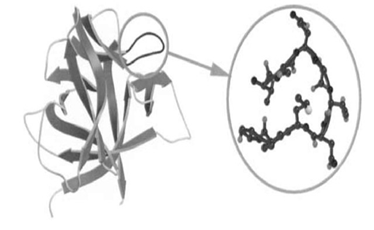
Figure 3: β-turn loop structure (A) and omega loop structure (B).
2.4 Coil structure
Coil structures are not true secondary structure but they mostly classified as the coil conformations. Coils are mostly located in a protein at places where amino acid residues do not form regular secondary structure such as α-helix or β-pleated sheet.
Though they are not regular structure lacking repetitive order but still they form stable conformations. Coil structure also has disordered regions which are called random coil structure. These structures also play important roles in protein function such as they can recognize ligand and help in their binding to the protein.
References
- Eisenberg, D. (2003). The discovery of the α-helix and β-sheet, the principal structural features of proteins. Proceedings of the National Academy of Sciences, 100(20), 11207-11210.
- Yang, Y., Gao, J., Wang, J., Heffernan, R., Hanson, J., Paliwal, K., & Zhou, Y. (2016). Sixty-five years of the long march in protein secondary structure prediction: the final stretch?. Briefings in bioinformatics, 19(3), 482-494.
- Jeong, W. H., Lee, H., Song, D. H., Eom, J. H., Kim, S. C., Lee, H. S., … & Lee, J. O. (2016). Connecting two proteins using a fusion alpha helix stabilized by a chemical cross linker. Nature communications, 7, 11031.
- Zhang, C., & Kim, S. H. (2000). The anatomy of protein β-sheet topology. Journal of molecular biology, 299(4), 1075-1089.
- Shapovalov, M., Vucetic, S., & Dunbrack Jr, R. L. (2019). A new clustering and nomenclature for beta turns derived from high-resolution protein structures. PLoS computational biology, 15(3), e1006844.
- Singh, M. (2006). Predicting protein secondary and supersecondary structure.
- Perskie, L. L., & Rose, G. D. (2010). Physical–chemical determinants of coil conformations in globular proteins. Protein Science, 19(6), 1127-1136.